Hunger is not a hindrance for N-terminal acetylation
Arnesen Lab reveals that the global level of N-terminal acetylation, a highly abundant protein modification in eukaryotes, remains stable in starving yeast cells despite an overall decrease in the cellular level of acetyl-CoA. The findings, described in the December issue of Molecular & Cellular Proteomics¸ represent the first comprehensive study of metabolic regulation of N-terminal acetylation.

Main content
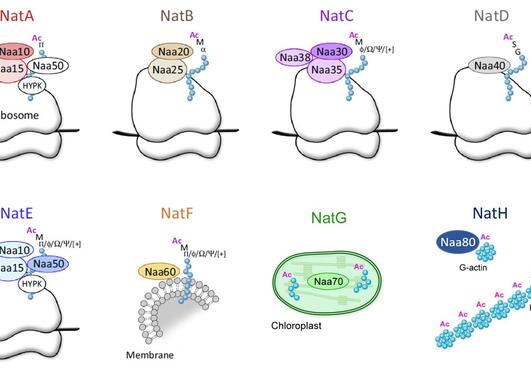
Our cells sense and respond to environmental cues. This fundamental process is essential for cell viability and function. Acetyl-CoA is a central metabolite and signaling molecule that play crucial roles in numerous cellular processes, including energy production and protein modification. Acetylation is one of the most common proteins modifications in eukaryotes and entails the addition of an acetyl moiety to the free α-amino group of protein N-termini (N-terminal acetylation) or to the ε-amino group of internal lysine residues (lysine acetylation). These acetylation reactions are catalyzed by N-terminal acetyltransferases (NATs) or lysine acetyltransferases (KATs), respectively. Acetyl-CoA serves as acetyl donor for all acetyltransferases, indicating a link between protein acetylation and cellular metabolism. Previous studies in different model organisms show that the global histone acetylation levels are sensitive to the concentration of acetyl-CoA in the cell (reviewed by Sivanand et al, 2018, TiBS). Could the same be the case for N-terminal acetylation and thus be a general adaptive concept in nature?
To address this issue, Dr. Sylvia Varland and Prof. Thomas Arnesen along with a team of other researchers investigated the effect of prolonged starvation on the global levels of N-terminal acetylation in the budding yeast Saccharomyces cerevisiae. Using a positional proteomic technique called N-terminal COFRADIC, the researchers found that yeast cells maintain their global N-terminal acetylation levels upon prolonged nutrient starvation, despite a marked decrease in acetyl-CoA levels. Moreover, they observed two distinct set of protein N-termini that display differential and opposing N-terminal acetylation behavior following starvation, indicating a dynamic process. Intriguingly, one of the protein clusters showed increased N-terminal acetylation levels as the yeast cells progressed to stationary phase in response to nutrient starvation. This finding was somewhat unexpected seeing that histone acetylation follows the opposite trend, becoming less acetylated when the amount of acetyl-CoA is reduced. Specifically, they found that the degree of N-terminal acetylation of Pcl8, a negative regulator of glycogen biosynthesis and two components of the pre-ribosome complex (Rsa3 and Rpl7a) increased during starvation. Furthermore, the steady-state levels of these proteins were regulated both by starvation and NatA activity. The authors suggest that specific, rather than global, N-terminal acetylation events are subject to metabolic regulation.
“Our study is the first investigation of a potential metabolic regulation of N-terminal acetylation,” says Dr. Sylvia Varland who is a researcher at the Department of Biological Sciences and the Department of Biomedicine at University of Bergen and currently on a research stay at the Donnelly Center at University of Toronto in Canada. “Our investigations demonstrates that N-terminal acetylation is a very robust protein modification. We think that regulation of N-terminal acetylation levels of certain subset of proteins are critical for the cell to adapt to enviromental cues”.
The study was made possible due to collaboration between the teams of Prof. Thomas Arnesen and Prof. Mathias Ziegler at University of Bergen, Norway and the Gevaert lab at Ghent University, Belgium.
This work was supported by research grants from the Research Council of Norway (NFR), Bergen Research Foundation (BFS), Norwegian Cancer Society, the Western Norway Regional Health Authority (WNRHA, Helse Vest), Meltzer Research Fund, and the Research Foundation - Flanders (FWO).